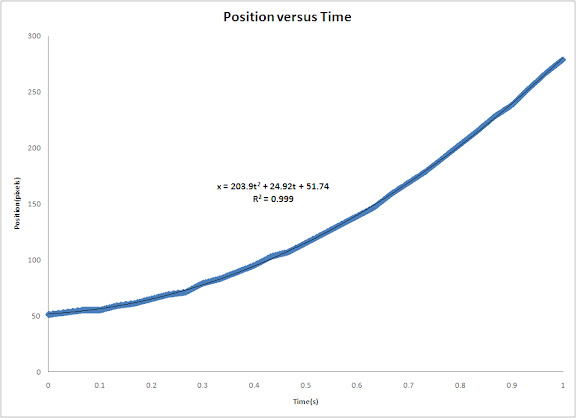
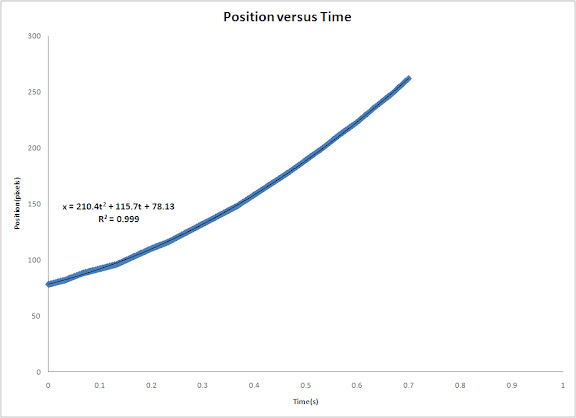
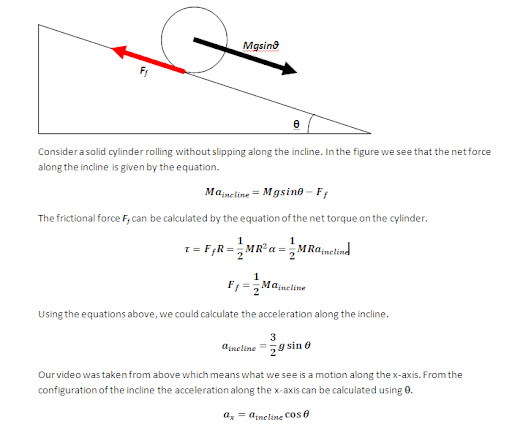
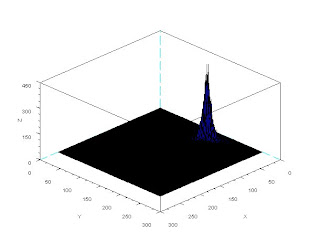
In this activity we would try to classify a set of images using the minimum distance classification.
Let wj where j = 1, 2, 3 ... W be a set of classes and W the total number of classes. If we define a representative of class wj to be its mean feature vector then
where xj is the set of ALL feature vectors in class wj and Nj is the number of samples in class wj. The simplest way of determining class membership is to classify an unknown feature vector x to the class whose mean or representative it is nearest to. For example, we can use the Euclidean distance
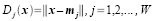
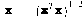
The objects I tried to sort out are the foods we bought near the math building. These are quail eggs, squid balls, chicken balls, fish balls, v-cut, piatos, and pillows. (We almost ate it while we were on the way back to CSRC.lol) We took images of each of these foods by their class then obtained their feature vector. The feature vector I used contained the mean chromaticity values r and g of the class as well as the mean of the standard deviation of its chromaticity. I believe these are the information that would most distinguish each class from one another. The images are shown below (drools....)














I tried to classify 21 samples using the minimum distance classification method and got the follow results
The accuracy of classification is 90%. This is quite good. This means the classifiers we used are indeed the relevant information.
I give myself a grade of 10 out of 10 for this activity for having classified samples with a fairly high accuracy. I thank Raf, Jorge, Ed and Billy for helping me in this activity. Special thanks to Cole, JC and Benj for the food.^_^
| Predicted | ||||||||||
| quail eggs | squid balls | chicken balls | fish balls | piatos | v-cut | pillows | ||||
| Actual | quail eggs | 3 | ||||||||
| squid balls | 3 | |||||||||
| chicken balls | 3 | |||||||||
| fish balls | 3 | |||||||||
| piatos | 3 | |||||||||
| v-cut | 1 | 1 | 1 | |||||||
| pillows | 3 | |||||||||
The accuracy of classification is 90%. This is quite good. This means the classifiers we used are indeed the relevant information.
I give myself a grade of 10 out of 10 for this activity for having classified samples with a fairly high accuracy. I thank Raf, Jorge, Ed and Billy for helping me in this activity. Special thanks to Cole, JC and Benj for the food.^_^